Software
DataLad

DataLad is ongoing work funded by NSF and German BMBF, to adapt the model of open-source software (OSS) distributions to address the technical limitations of today's data-sharing and provides a versatile data management platform. It uses software for data tracking and deployment logistics specialized for large data (git-annex) built atop Git, the most capable distributed version control system (dVCS) available today. DataLad provides access to data available from various sources (e.g. lab or consortium web-sites such as humanconnectome.org; data sharing portals such as openneuro.org and crcns.org) through a single interface. It enables students and scientists to operate on data using familiar concepts, such as files and directories, while transparently managing data access and authorization with underlying hosting providers.
- M. Hanke, M. Visconti di Oleggio Castello, K. Meyer, B. Poldrack, and Y.O. Halchenko (2018). YODA: YODA's organigram on data analysis. OHBM 2018, Singapore.
- JOSS paper: providing a succinct overview of DataLad
- DataLad Handbook: everything you need to know about DataLad.
- Funding support
DANDI

Distributed Archives for Neurophysiology Data Integration (DANDI) is a platform for publishing, sharing, and processing neurophysiology data funded by the BRAIN Initiative. The platform is now available for data upload and distribution, and provides supplementary client tools to assist with introspection and organization of data following NWB standard.
- NIH (#1R24MH117295-01A1). PIs: S. Ghosh and Y.O. Halchenko
EMBER

Ecosystem for Multi-modal Brain-behavior Experimentation and Research (EMBER) is the BRAIN Initiative archive for multi-modal neurophysiological and behavioral data, supporting the Brain Behavior Quantification and Synchronization (BBQS) Program. The platform is being kick-started based on DANDI to provide data upload and distribution. CON is working on integration with DANDI and involved standards.
- NIH (#1R24MH136632-01 ). PI: B.A. WESTER, Co-I: Y.O. Halchenko
OpenNeuro

OpenNeuro is likely the oldest and the most well known BRAIN Initiative archive for neuroimaging data. One of the exciting features of the project is that being inspired by our early DataLad crawling of its OpenfMRI.org precursor, it is now using git-annex directly as its core technology. CON is contributing in ensuring availability of recent versions of git-annex and DataLad, involved standardization efforts, and inter-archive collaborations.
- NIH (#5R24MH117179-07). PI: R. Poldrack, Co-I: Y.O. Halchenko
DataLad-Registry

DataLad-Registry is a service that maintains up-to-date information on an expanding collection of datasets, currently numbering over ten thousand. It automatically registers datasets from various online sources, extracts metadata, and keeps both the datasets and their metadata current. DataLad-Registry offers search functionalities, including metadata-based searches, accessible through both a web interface and a RESTful API. Registry is regularly updated with DataLad datasets discovered across multiple hosting portals and listed on our DataLad Usage Dashboard.
-
Talk at distribits 2024: Bringing Benefits of Centrality to Datalad (click to expand)
DueCredit

DueCredit provides solution for the problem of inadequate citation and referencing of scientific software and methods. It provides a simple framework (at the moment for Python only) to embed publication or other references in the original code so they are automatically collected and reported to the user at the necessary level of reference detail, i.e. only references for actually used functionality will be presented back if software provides multiple citeable implementations.
As a side-effect, we hope that DueCredit also will reduce demand in "prima-ballerina" projects, will encourage contributions to existing open-source codebases, and as a result would solidify scientific software ecosystem.
- Y.O. Halchenko and M. Visconti di Oleggio Castello (2016). DueCredit - automagically collect citations for software, methods, and data you use. OHBM 2016, Geneva, Switzerland
HeuDiConv / ReproIn
HeuDiConv is a flexible DICOM converter for organizing brain imaging data into structured directory layouts. As a part of the larger, NIH supported ReproNim effort, we are developing a HeuDiConv-based ReproIn solution for turnkey automatic conversion of all collected MR data to a collection of the BIDS DataLad datasets. It includes a flexible BIDS-like specification how to name scanning sequences in the scanner, and a HeuDiConv reproin.py heuristic to automate layout and conversion of the datasets. This solution is deployed at DBIC (Dartmouth Brain Imaging Center), it already facilitates reproducible research, data sharing, and uploads to central archives such as NDA.
- Halchenko et al., (2024). HeuDiConv — flexible DICOM conversion into structured directory layouts. Journal of Open Source Software, 9(99), 5839 DOI: 10.21105/joss.05839.
- M. Visconti di Oleggio Castello, James E. Dobson, Terry Sackett, Chandana Kodiweera, J.V. Haxby, M. Goncalves, S. Ghosh, Y.O. Halchenko ReproIn: automatic generation of shareable, version-controlled BIDS datasets from MR scanners, OHBM 2018, Singapore.
NeuroDebian

NeuroDebian is a turnkey research software platform for all aspects of the neuroscientific research process. It takes the ideas of the software hosting portals such as NITRC on maximizing research transparency and methods sharing, one step further, by providing a comprehensive suite of readily usable and fully integrated software with a robust testing and deployment infrastructure. Consequently, it improves interoperability among the tools and frees researchers from the burden of tedious installation or upgrade procedures. That, in turn, positively affects their availability for actual research activities, as well as their motivation to test new analysis tools and stay connected with the latest methodological developments in the field.
- Y.O. Halchenko & M. Hanke (2012). Open is not enough. Let's take the next step: An integrated, community-driven computing platform for neuroscience. Frontiers in Neuroinformatics, 6:22. [PDF] DOI: 10.3389/fninf.2012.00022
PyMVPA

PyMVPA is a Python-based framework for neural decoding using multivariate pattern analysis. It affords both volume- and surface-based analyses using a wide variety of supervised and unsupervised machine learning methods, representational similarity analyses, searchlight analyses, hyperalignment of representational spaces, and model-based decoding and encoding. The software also can be used for neural data other than fMRI, including analysis of MEG and EEG data through spatio-temporo-frequency band searchlights and cross-modal EEG to fMRI trans-fusion. It also has been used for analyses on data unrelated to neuroscience, demonstrating its general utility. PyMVPA also serves as a repository for sample data sets (e.g., Haxby et al. 2001) that has found wide applicability for education, development of new algorithms, or new analyses and independent research reports.
- M. Hanke, Y.O. Halchenko, et al. (2009). PyMVPA: A Python toolbox for multivariate pattern analysis of fMRI data. Neuroinformatics, 7, 37-53. DOI: 10.1007/s12021-008-9041-y
ReproMan

ReproMan (Reproducible computational environments Manager; formerly known as REPROMAN) is also a part of the NIH supported ReproNim effort. It aims to facilitate reproducible computation via collection of detailed information about origin of the used components (Debian and/or Conda packages, VCS repositories, etc), so that computational environments could be analyzed, and re-created.
- M. Travers, R. Buccigrossi, C. Haselgrove, K. Meyer, and Y.O. Halchenko NICEMAN: NeuroImaging Computational Environments Manager, OHBM 2018, Singapore.
con/duct

con/duct (or just duct) is a lightweight wrapper that collects execution data for an arbitrary command. Execution data includes execution time, system information, and resource usage statistics of the command and all its child processes. duct is intended to simplify the problem of recording the resources necessary to execute a command, particularly in an HPC environment. Additionally duct includes optional helpers that interact with logs produced by duct exectutions.
(con/)tinuous
tinuous is a command for downloading build logs and (for GitHub only) artifacts and release assets for a GitHub repository from GitHub Actions, Travis-CI.com, and/or Appveyor. By downloading them all, and optionally placing them under DataLad control you can establish the backup, distribution, and convenient harmonious access to all those artifacts.
pyout

pyout is a Python package that defines an interface for writing structured records as a table in a terminal. It is being developed to replace custom code for displaying tabular data in in DANDI client and others.
Quail

Quail is a Python toolbox for analyzing data from free recall memory experiments. Some key features include:
- A simple data structure for storing encoding and recall data
- A set of functions for analyzing data by computing standard memory performance metrics
- A simple API for customizing plot styles
- Support for "naturalistic" stimuli such as movies, texts, and speech data
- A set of powerful tools for importing data, automatically transcribing audio files (speech-to-text), and more
- A.C. Heusser, P.C. Fitzpatrick, C.E. Field, K. Ziman, and J.R. Manning (2017). Quail: A Python toolbox for analyzing and plotting free recall data. The Journal of Open Source Software, 2(18): 424.
HyperTools

HyperTools is a Python toolbox for gaining geometric insights into high dimensional data. Features include:
- Functions for plotting high-dimensional datasets in 2D and 3D
- Static and animated plots
- Simple API for customizing plot styles
- Set of powerful data manipulation tools including hyperalignment, k-means clustering, normalizing, and more
- Support of lists of Numpy arrays, Pandas dataframes, text, or (mixed) lists
- Applying topic models and other text and word embedding methods to text data
- A.C. Heusser, K. Ziman, L.L.W. Owen, and J.R. Manning (2018). HyperTools: a Python Toolbox for Gaining Geometric Insights into High-Dimensional Data. Journal of Machine Learning Research, 18: 1-6.
SuperEEG

SuperEEG is a Python toolbox for inferring whole-brain activity from sparse ECoG recordings. The way the technique works is to leverage data from different patients' brains (who had electrodes implanted in different locations) to learn a "correlation model" that describes how activity patterns at different locations throughout the brain relate. Given this model, along with data from a sparse set of locations, we use Gaussian process regression to "fill in" what the patients' brains were "most probably" doing when those recordings were taken. Details on our approach may be found in this preprint. You may also be interested in watching this talk or reading this blog post from a recent conference.
- L.L.W. Owen, A.C. Heusser, and J.R. Manning (2018). A Gaussian process model of human electrocorticographic data. bioRxiv, 121020.
Initiatives
Open Brain Consent

Open Brain Consent initiative aims to facilitate neuroimaging data sharing by providing an "out of the box" solution addressing aforementioned human subjects concerns and consisting of
- widely acceptable consent form allowing deposition of anonymized data to public data archives
- collection of tools/pipelines to help anonymization of neuroimaging data making it ready for sharing
Initiated and managed by CON, OBC is now an international effort with materials for GDPR compliance and multiple internationalization efforts to make sure materials are available in all applicable languages. Contribute if you see your language is missing!
-
Bannier E, Barker G, Borghesani V, Broeckx N, Clement P,
Emblem KE, Ghosh S, Glerean E, Gorgolewski KJ, Havu
M, Halchenko YO, Herholz P, Hespel A, Heunis
S, Hu Y, Hu CP, Huijser D, de la Iglesia Vayá M, Jancalek R,
Katsaros VK, Kieseler ML, Maumet C, Moreau CA, Mutsaerts HJ,
Oostenveld R, Ozturk-Isik E, Pascual Leone Espinosa N, Pellman
J, Pernet CR, Pizzini FB, Trbalić AŠ, Toussaint PJ, Visconti
di Oleggio Castello M, Wang F, Wang C, Zhu H.
The Open Brain Consent: Informing research participants and obtaining consent to share brain imaging data . Hum Brain Mapp. 2021 May;42(7):1945-1951. doi: 10.1002/hbm.25351. Epub 2021 Feb 1. PMID: 33522661; PMCID: PMC8046140.
Meetings
For over a decade, CON and its projects are presented annually with exhibits at the annual meetings of the Organization for Human Brain Mapping (OHBM) Society and the Society for Neuroscience. In collaboration with Soichi Hayashi and other wonderful people we proudly saved the first ever virtual OHBM 2020 by providing a virtual posters venue.distribits

distribits is a community for enthusiasts of tools and workflows in the domain of distributed data. Take a look at the topics of past meeting contributions, and YouTube videos from distribits 2024 to learn what is being talked about at distribits.
Standards
Brain Imaging Data Structure (BIDS)
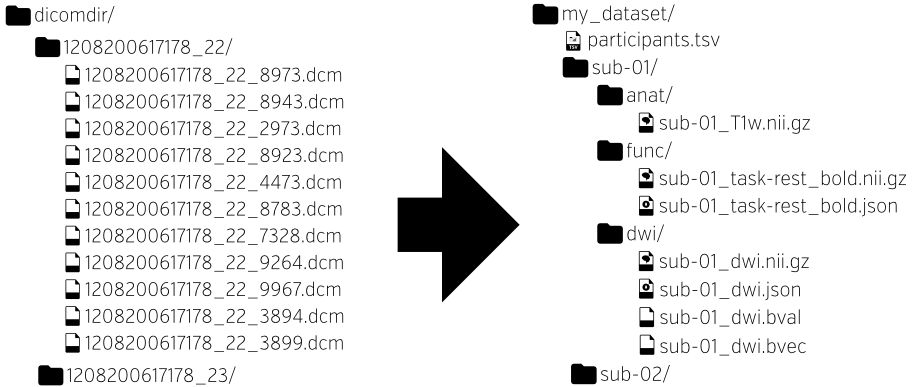
BIDS is a project lead by a steering group elected by the BIDS community to provide a simple and intuitive way to organize and describe your neuroimaging and behavioral data.
- Gorgolewski, K. J., et many, Y.O. Halchenko et many more (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3. DOI: 10.1038/sdata.2016.44
- Example (test) datasets
- Historical OpenfMRI and up-to-date OpenNeuro Datasets in DataLad distribution.
Neurodata Without Borders: Neurophysiology (NWB:N)

NWB:N is a data standard for neurophysiology, providing neuroscientists with a common standard to share, archive, use, and build analysis tools for neurophysiology data. It is a standard supported by BIDS and the DANDI archive.
YODA's Organigram on Data Analysis

YODA is a collection of best practices for organizing and managing digital objects of science.
- Michael Hanke, Kyle A. Meyer, Matteo Visconti di Oleggio Castello, Benjamin Poldrack, Yaroslav O. Halchenko. (2018) YODA: YODA's Organigram on Data Analysis F1000Research, 7:1965 (poster) DOI 10.7490/f1000research.1116363.1
Education
ReproNim: Reproducible Basics
Reproducible Basics training module of the ReproNim training curriculum presents daily core tools (shell, version control, etc) and explains how you could make your research more reproducible having gained improved knowledge of them.
- Y.O. Halchenko et al.
Infrastructure
SingularityHub (after-life)
To provide archival and uninterrupted access to over 9TBs of singularity-hub.org Singularity containers, we have established ///shub DataLad dataset and a service to serve all the shub:// URLs.